What We Do
The Kretz Lab is focused on understanding the biochemical and genetic regulation of the blood clotting system. We use traditional tools in protein biochemistry and molecular biology to map protein interactions and define the regulation of proteases in the cardiovascular system. We also employ high throughput screening techniques to understand the impact of genetic variants on blood clotting genes. Our goal is to develop novel tools to treat and diagnose cardiovascular diseases. We collaborate extensively with researchers at TaARI, McMaster University and beyond to achieve our research goals.
Who We Are
Dr. Colin Kretz is an associate professor in the Department of Medicine, division of hematology and thromboembolism. Dr. Kretz grew up in Dundas Ontario and completed his PhD at McMaster University under the supervision of Dr. Jeffrey Weitz, from 2004-09. During that time, he formed a foundation in protein biochemistry studying the serine protease thrombin. He then moved to the University of Michigan for postdoctoral training in the lab of David Ginsburg, from 2009-16. There, Dr. Kretz developed expertise in human and mouse genetics to complement his training in protein biochemistry. Dr. Kretz joined McMaster University and TaARI as an assistant professor in 2016.
Current Lab Members
Colin Kretz
PhD
Associate Professor,
Department of Medicine

Colin Kretz
PhD
Associate Professor,
Department of Medicine
Taylor Sparring
PhD Student
Project: Deep Mutational Scanning of the VWF C domains
Peter Andrisani
MSc Student
Project: Exploring the mechanism of ADAMTS13 latency
Rex Huang
MSc Student
Project: Developing novel ADAMTS13 variants as therapeutics
Colin Kretz
PhD
Associate Professor,
Department of Medicine
Colin Kretz
PhD
Associate Professor,
Department of Medicine
Taylor Sparring
PhD Student
Project: Deep Mutational Scanning of the VWF C domains
Taylor Sparring
PhD Student
Project: Deep Mutational Scanning of the VWF C domains
Peter Andrisani
MSc Student
Project: Exploring the mechanism of ADAMTS13 latency
Peter Andrisani
MSc Student
Project: Exploring the mechanism of ADAMTS13 latency
Rex Huang
MSc Student
Project: Developing novel ADAMTS13 variants as therapeutics
Rex Huang
MSc Student
Project: Developing novel ADAMTS13 variants as therapeutics
Former Students
Veronica DeYoung
MSc Student
Thesis: Characterizing protease resistant mutants of ADAMTS13
Current Position: Medical Student, Queens University
Veronica DeYoung
MSc Student
Kanwal Singh
PhD Student
Thesis: ADAMTS13 Resistance to Protease Inhibition
Current Position: Postdoctoral Fellow, St. Michael’s Hospital
Kanwal Singh
PhD Student
Veronica DeYoung
MSc Student
Thesis: Characterizing protease resistant mutants of ADAMTS13
Current Position: Medical Student, Queens University
Veronica DeYoung
MSc Student
Thesis: Characterizing protease resistant mutants of ADAMTS13
Current Position: Medical Student, Queens University
Kanwal Singh
PhD Student
Thesis: ADAMTS13 Resistance to Protease Inhibition
Current Position: Postdoctoral Fellow, St. Michael’s Hospital
Kanwal Singh
PhD Student
Thesis: ADAMTS13 Resistance to Protease Inhibition
Current Position: Postdoctoral Fellow, St. Michael’s Hospital
Projects
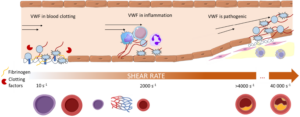
Current research projects are primarily focused on understanding the regulation of the VWF/ADAMTS13 axis, which is important at the earliest phases of blood clotting. Von Willebrand Factor (VWF) is a long string-like protein that adheres to the site of vessel injury and captures platelets flowing in blood. Recruitment of too many platelets can lead to thrombosis. ADAMTS13 is a protease that cuts VWF strings, shortening them and reducing their platelet-capturing activity. Dysregulation of VWF and ADAMTS13 can lead to an increased risk of thrombosis in both chronic diseases (TTP, obesity, diabetes, atherosclerosis) and acute illnesses (sepsis, trauma). Understanding the mechanisms of their dysregulation may lead to the development of novel therapies to treat thrombosis.
Expandable List
The objective of this project is to explore the biochemical and structural mechanisms that contribute to the regulation of ADAMTS13. ADAMTS13 exists as a latent protease that undergoes substrate-induced activation through a mechanism that is not well understood. Because ADAMTS13 is an important regulator of blood clotting, understanding the mechanisms that control its activity will allow us to design novel therapies to treat thrombosis.
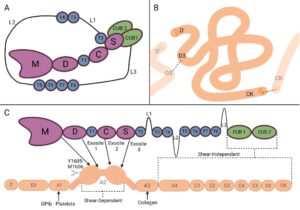
Figure: VWF recognition by ADAMTS13. (A) ADAMTS13 circulates in a closed conformation that masks important exosites required for binding to VWF. (B) VWF is a large multimeric protein that circulates in a compact “birds nest” configuration. (C) Under conditions of high shear stress (such as at the site of vessel injury), VWF unfolds into a long string like form, exposing critical binding sites for ADAMTS13. As the VWF multimer accumulates platelets, its central A2 domain can denature to expose the cleavage site for ADAMTS13.
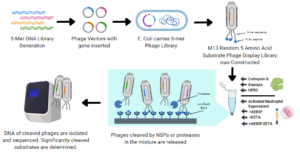 Approximately two per cent of the human genome encodes for a protease. Many of these proteases are poorly understood, having no known role in biology. Therefore, new tools are required to explore the role of proteases in biological processes. We are developing and applying a high throughput technique to define protease substrates specificity, termed deep protease profiling. This technique is based on the coupling of traditional substrate phage display with high throughput next generation DNA sequencing. This approach allows us to quantitatively determine the activity of a protease towards >3.2 million peptide sequences in a single reaction and then use computational methods to understand the rules that govern its substrate specificity. The long-term goals of this project are to develop a tool kit of substrates with absolute specificity toward its target protease and to identify protease activity in complex biological mixtures. Such technologies will shed new light on the role of proteases in biology and may lead to the develop of novel diagnostic tools to detect protease activity in biological settings.
Approximately two per cent of the human genome encodes for a protease. Many of these proteases are poorly understood, having no known role in biology. Therefore, new tools are required to explore the role of proteases in biological processes. We are developing and applying a high throughput technique to define protease substrates specificity, termed deep protease profiling. This technique is based on the coupling of traditional substrate phage display with high throughput next generation DNA sequencing. This approach allows us to quantitatively determine the activity of a protease towards >3.2 million peptide sequences in a single reaction and then use computational methods to understand the rules that govern its substrate specificity. The long-term goals of this project are to develop a tool kit of substrates with absolute specificity toward its target protease and to identify protease activity in complex biological mixtures. Such technologies will shed new light on the role of proteases in biology and may lead to the develop of novel diagnostic tools to detect protease activity in biological settings.
Mutations in VWF that impair its clotting function result in the bleeding disorder von Willebrand Disease. While genome sequencing efforts identify genetic variants in VWF, the major obstacle is in the interpretation of variants. This is due to a lack of evidence about the functional impact of variants. Traditional phenotyping efforts suffer from low throughput and cannot keep up with the demand. The result is a growing list of variants of unknown significance (VUS). In fact, approximately 41 per cent of the 72,000 missense variants annotated in the ClinVar database are classified as VUS. In this project, we are applying Deep mutational scanning (DMS) in order to catalogue the impact of every possible single amino acid substitution at each position of VWF on its function. To do this, we have developed a cell secretion assay in order to quantify the impact of missense mutations on VWF biosynthesis and secretion from HEK293 cells.
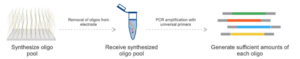
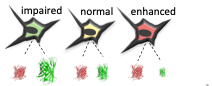
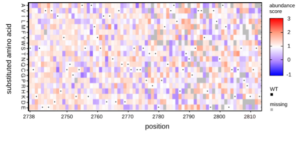
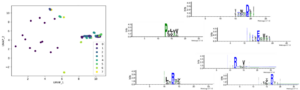
Figure: Mutagenesis libraries are designed in silico and synthesized as an oligonucleotide pool. Pooled oligo libraries are assembled into full length VWF expression vector, fused to eGFP. Libraries are stably transfected into HEK293 cells, and sorted into bins according to their intracellular eGFP signal. Bins are submitted for throughput DNA sequencing to quantify variant abundance within each bin. Data are expressed as heat maps representing cell abundance scores for every variants at each position.